Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Hirato, Misaki*; Yokoya, Akinari*; Baba, Yuji*; Mori, Seiji*; Fujii, Kentaro*; Wada, Shinichi*; Izumi, Yudai*; Haga, Yoshinori
Physical Chemistry Chemical Physics, 25(21), p.14836 - 14847, 2023/05
Times Cited Count:2 Percentile:77.93(Chemistry, Physical)Shimada, Mikio*; Tokumiya, Takumi*; Miyake, Tomoko*; Tsukada, Kaima*; Kanzaki, Norie; Yanagihara, Hiromi*; Kobayashi, Junya*; Matsumoto, Yoshihisa*
Journal of Radiation Research (Internet), 64(2), p.345 - 351, 2023/03
Times Cited Count:0 Percentile:0.01(Biology) -H2AX focus formation assay based on track-structure simulation
-H2AX focus formation assay based on track-structure simulationYachi, Yoshie*; Matsuya, Yusuke*; Yoshii, Yuji*; Fukunaga, Hisanori*; Date, Hiroyuki*; Kai, Takeshi
International Journal of Molecular Sciences (Internet), 24(2), p.1386_1 - 1386_14, 2023/01
Times Cited Count:2 Percentile:75.46(Biochemistry & Molecular Biology)When living cells are irradiated with radiation and complex damage is formed within a few nanometers of DNA, it is believed to induce biological effects such as cell death. In general, complex DNA damage formed in cells can be detected experimentally by fluorescence microscopy, because the area around the damage site emits light like a focus point when a fluorophore is used. However, this detection method has not been able to analyze the degree of complexity of DNA damage. Therefore, in this study, we addressed on the measured focus size and evaluated the degree of complexity of DNA damage using a track structure analysis code. As a result, we found that as DNA damage becomes more complex, the focus size also increases. Our findings are expected to provide a new analytical method for elucidating the initial factors of radiation biological effects.
Yachi, Yoshie*; Kai, Takeshi; Matsuya, Yusuke; Hirata, Yuho; Yoshii, Yuji*; Date, Hiroyuki*
Scientific Reports (Internet), 12, p.16412_1 - 16412_8, 2022/09
Times Cited Count:2 Percentile:47.19(Multidisciplinary Sciences)Recently, magnetic resonance-guided radiotherapy (MRgRT) which can visualize tumors in real time has been developed and installed in several clinical facilities. It is known that Lorentz force modulate macroscopic dose distribution by a charged particle, however, the impact by the force on microscopic radiation-track structure and early DNA damage induction remain unclear. In this study, we simulated the electron-track structure in a static magnetic field using a PHITS, and estimated features of biological effects. We indicated that the macroscopic dose distributions are changed by the force, while early DNA damage such as double strand breaks is attributed to the secondary electrons below a few tens of eV which are independent of the force. We expect that our insight significantly contributes to the MRgRT.
Kataoka, Takahiro*; Shuto, Hina*; Naoe, Shota*; Yano, Junki*; Kanzaki, Norie; Sakoda, Akihiro; Tanaka, Hiroshi; Hanamoto, Katsumi*; Mitsunobu, Fumihiro*; Terato, Hiroaki*; et al.
Journal of Radiation Research (Internet), 62(5), p.861 - 867, 2021/09
Times Cited Count:5 Percentile:55.27(Biology)Matsuura, Masato*; Yamada, Takeshi*; Tominaga, Taiki*; Kobayashi, Makoto*; Nakagawa, Hiroshi; Kawakita, Yukinobu
JPS Conference Proceedings (Internet), 33, p.011068_1 - 011068_6, 2021/03
The position dependence of the scattered intensity in the time-of-flight backscattering spectrometer DNA was investigated. A periodic structure for both vertical (pixel) and horizontal (PSD) directions was observed. The solar slit and over-bending of an analyzer crystal is discussed as a possible origin of the modulation in the intensity. We have developed software program for the systematic correction of the position-dependent intensity and offset energy for the elastic peak. This corrects the deviation from the true scattering intensity and improve the quality of the data, which includes the energy resolution.
Saga, Ryo*; Matsuya, Yusuke; Takahashi, Rei*; Hasegawa, Kazuki*; Date, Hiroyuki*; Hosokawa, Yoichiro*
Journal of Radiation Research, 60(3), p.298 - 307, 2019/05
Times Cited Count:23 Percentile:81.57(Biology)In radiotherapy, it is recognized that cancer stem cells (CSCs) in tumor tissue shows radio-resistance. However, the relationship between content percentage of the CSCs and dose-response curve on cell survival remain unclear. In this study, we developed a stochastic model considering progeny cells and stem cells, and investigated the impact of stem cells on radio-sensitivity. From the flow-cytometric analysis (cell experiments), the content percentage of stem cells was 3.2% or less which agreed well with the model estimation from the cell survival curve. Based on the verification, it is suggested that cell survival in high-dose range is largely affected by the CSCs. In addition, regarding the sub-population of stem cells, the present model well reproduces the dose response on lethal lesions to DNA comparing with the conventional LQ model. This outcome indicates that the stem cells must be considered for describing the dose-response curve in wide dose range.
Kai, Takeshi; Yokoya, Akinari*; Fujii, Kentaro*; Watanabe, Ritsuko*
Hoshasen Kagaku (Internet), (106), p.21 - 29, 2018/11
It is thought to that the biological effects such as cell death or mutation are induced by complex DNA damage which are formed by several damage sites within a few nm. As the prediction of complex DNA damage at an electron track end, we report our outcomes. These results indicate that DNA damage sites comprising multiple nucleobase lesions with a single strand breaks can be formed by multiple collisions of the electrons within 1 nm. This multiple damage site cannot be processed by base excision repair enzymes. Pre-hydrated electrons can also be produced resulting in an additional base lesion over a few nm from the multi-damage site. This clustered damage site may be finally converted into a double strand break. These DSBs include another base lesion(s) at their termini that escape from the base excision process and which may result in biological effect. Our simulation is useful to reveal phenomena involved in radiation physico-chemistry as well as the DNA damage prediction.
Kai, Takeshi; Yokoya, Akinari*; Ukai, Masatoshi*; Fujii, Kentaro*; Toigawa, Tomohiro; Watanabe, Ritsuko*
Physical Chemistry Chemical Physics, 20(4), p.2838 - 2844, 2018/01
Times Cited Count:22 Percentile:75.15(Chemistry, Physical)It is thought that complex DNA damage which induces in radiation biological effects is formed at radiation track end. Thus, the earliest stage of water radiolysis at the electron track end was studied to predict DNA damage. These results indicate that DNA damage sites comprising multiple nucleobase lesions with a single strand breaks can therefore be formed by multiple collisions of the electrons within three base pairs (3bp) of a DNA strand. This multiple damage site cannot be processed by base excision repair enzymes. However, pre-hydrated electrons can also be produced resulting in an additional base lesion more than 3bp away from the multi-damage site. This clustered damage site may be finally converted into a double strand break (DSB) when base excision enzymes process the additional base lesions. These DSBs include another base lesion(s) at their termini that escape from the base excision process and which may result in biological effects such as mutation in surviving cells.
Watanabe, Ritsuko*; Kai, Takeshi; Hattori, Yuya*
Radioisotopes, 66(11), p.525 - 530, 2017/11
To understand the mechanisms of radiation biological effects, modeling and simulation studies are important. In particular, simulation approach is powerful tool to evaluate modeling of mechanisms and the relationship among experimental results in different spatial scale of biological systems such as DNA molecular and cell. This article summarizes our approach to evaluate radiation action on DNA and cells by combination of knowledge in radiation physics, chemistry and biology. It contains newly theoretical approach to estimate physico-chemical process of DNA damage induction in addition to typical method of DNA damage prediction. Outline of the mathematical model for dynamics of DNA damage and cellular response is also presented.
Hata, Kuniki; Lin, M.*; Yokoya, Akinari*; Fujii, Kentaro*; Yamashita, Shinichi*; Muroya, Yusa*; Katsumura, Yosuke*
Hoshasen Kagaku (Internet), (103), p.29 - 34, 2017/04
Reactivity of edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one), which is known to show high antioxidative properties, with oxidative species, such as hydroxyl radical (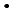 OH) and azide radical (N
OH) and azide radical (N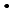
 ), was investigated by a pulse radiolysis experiment, and generation behavior of edaravone radicals produced through these reactions were observed. It was shown that OH-adducts are produced by the reaction with
), was investigated by a pulse radiolysis experiment, and generation behavior of edaravone radicals produced through these reactions were observed. It was shown that OH-adducts are produced by the reaction with 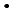 OH in contrast to the other oxidative radicals, which react with edaravone by an electron transfer reaction. Chemical repair properties of edaravone against DNA lesions produced by reactions of DNA with oxidative species were also investigated by a pulse radiolysis experiment with deoxyguanosine monophosphate (dGMP) and a
OH in contrast to the other oxidative radicals, which react with edaravone by an electron transfer reaction. Chemical repair properties of edaravone against DNA lesions produced by reactions of DNA with oxidative species were also investigated by a pulse radiolysis experiment with deoxyguanosine monophosphate (dGMP) and a  -radiolysis experiment with plasmid DNA solutions. It was observed that edaravone reduced dGMP radicals just after produced in a dilute aqueous solution and inhibited some base lesions on plasmid DNA more effectively than single strand breaks. These results show that edaravone may protect living system from oxidative stress, such as radiation, by not only scavenging oxidative species but also reducing precursors of DNA leisons.
-radiolysis experiment with plasmid DNA solutions. It was observed that edaravone reduced dGMP radicals just after produced in a dilute aqueous solution and inhibited some base lesions on plasmid DNA more effectively than single strand breaks. These results show that edaravone may protect living system from oxidative stress, such as radiation, by not only scavenging oxidative species but also reducing precursors of DNA leisons.
Kai, Takeshi; Yokoya, Akinari*; Fujii, Kentaro*; Watanabe, Ritsuko*
Yodenshi Kagaku, (8), p.11 - 17, 2017/03
It is thought to that the biological effects such as cell death or mutation are induced by complex DNA damage which are formed by several damage sites within a few nm. We calculated dynamic behavior of secondary electrons produced by primary electron and positon of high energy in water whose composition ratio is similar to biological context. The secondary electrons induce the ionization or electronic excitation near the parent cations. The decelerated electrons about 10% are distributed to their parent cations by the attractive Coulombic force. From the results, we predicted the following formation mechanism for the complex DNA damage. The electrons ejected from DNA could induce the ionization or the electronic excitation within the DNA. The electrons attracted by the Coulombic force are pre-hydrated in water layer of the DNA. The pre-hydrated electrons could induce to the DNA damage by dissociative electron transfer. As the results, the complex DNA damage with 1 nm could be formed by the interaction of not only the primary electron or positon but also the secondary electrons.
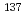 Cs
CsWatanabe, Ritsuko; Hattori, Yuya; Kai, Takeshi
International Journal of Radiation Biology, 92(11), p.660 - 664, 2016/11
Times Cited Count:2 Percentile:19.71(Biology)To understand the effect of internal exposure of 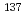 Cs, we focus on estimation of microscopic energy deposition pattern and DNA damage induced by directly emitted electrons (beta-rays, internal conversion electrons, Auger electrons) from
Cs, we focus on estimation of microscopic energy deposition pattern and DNA damage induced by directly emitted electrons (beta-rays, internal conversion electrons, Auger electrons) from 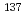 Cs. Monte Carlo track simulation method was used to calculate the microscopic energy deposition pattern. To simulate the energy deposition by directly emitted electrons, we considered the multiple ejections of electrons after internal conversion. Induction process of DNA strand breaks and base lesions was modeled and simulated using Monte Carlo methods for cell mimetic condition. The yield and spatial distribution of simple and complex DNA damage were calculated for the cases of
Cs. Monte Carlo track simulation method was used to calculate the microscopic energy deposition pattern. To simulate the energy deposition by directly emitted electrons, we considered the multiple ejections of electrons after internal conversion. Induction process of DNA strand breaks and base lesions was modeled and simulated using Monte Carlo methods for cell mimetic condition. The yield and spatial distribution of simple and complex DNA damage were calculated for the cases of  -rays and electrons from
-rays and electrons from 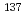 Cs. The simulation showed that significant difference of DNA damage spectrum was not caused by the difference between secondary electron spectrum by
Cs. The simulation showed that significant difference of DNA damage spectrum was not caused by the difference between secondary electron spectrum by  -rays and directly ejected electron spectrum. The result support that the existing evaluation that internal exposure and external exposure are almost equivalent.
-rays and directly ejected electron spectrum. The result support that the existing evaluation that internal exposure and external exposure are almost equivalent.
Kai, Takeshi; Yokoya, Akinari; Ukai, Masatoshi; Fujii, Kentaro; Watanabe, Ritsuko
International Journal of Radiation Biology, 92(11), p.654 - 659, 2016/11
Times Cited Count:10 Percentile:68.36(Biology) is characterized as Type II bacterial topoisomerase and its activity is differentially regulated by PprA in vitro
is characterized as Type II bacterial topoisomerase and its activity is differentially regulated by PprA in vitroKota, S.*; Rajpurohit, Y. S.*; Charaka, V. K.*; Sato, Katsuya; Narumi, Issey*; Misra, H, S.*
Extremophiles, 20(2), p.195 - 205, 2016/03
Times Cited Count:12 Percentile:40.92(Biochemistry & Molecular Biology)Izumi, Yudai; Yamamoto, Satoshi*; Fujii, Kentaro; Yokoya, Akinari
Hoshasen Seibutsu Kenkyu, 51(1), p.91 - 106, 2016/03
no abstracts in English
Ghobadi, A. F.*; Letteri, R.*; Parelkar, S. S.*; Zhao, Y.; Chan-Seng, D.*; Emrick, T.*; Jayaraman, A.*
Biomacromolecules, 17(2), p.546 - 557, 2016/02
Times Cited Count:15 Percentile:55.55(Biochemistry & Molecular Biology)Balestrazzi, A.*; Achary V Mohan Murali*; Macovei, A.*; Yoshiyama, Kaoru*; Sakamoto, Ayako
Frontiers in Plant Science (Internet), 7, p.64_1 - 64_2, 2016/02
Times Cited Count:3 Percentile:56.51(Plant Sciences)no abstracts in English
Kaburagi, Masaaki; Yamada, Hironao*; Miyakawa, Takeshi*; Morikawa, Ryota*; Takasu, Masako*; Kato, Takamitsu*; Uesaka, Mitsuru*
Polymer Journal, 48(2), p.189 - 195, 2016/02
Times Cited Count:5 Percentile:18.83(Polymer Science)We performed molecular dynamics (MD) simulations of telomeric single-stranded DNA and POT1 for 100 ns. The distance between 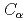 (POT1) and O5' (telomeric ssDNA) is calculated to verify the binding system for 100 ns MD. We then calculated the distance between the bases of telomeric DNA ends and the root mean square deviation and gyration radius in single and binding states. We compared the root mean square fluctuations between single and binding states and calculated the number of hydrogen bonds between POT1 and telomeric DNA. There are many hydrogen bonds between Gln94 and the first guanine of the closest TTAGGG sequence in telomeric single-stranded DNA. These Gln94 and the guanine have a large difference in root mean square fluctuation between single and binding states. We found that Gln94 and guanine are important components of the binding system, and they are related to its stability.
(POT1) and O5' (telomeric ssDNA) is calculated to verify the binding system for 100 ns MD. We then calculated the distance between the bases of telomeric DNA ends and the root mean square deviation and gyration radius in single and binding states. We compared the root mean square fluctuations between single and binding states and calculated the number of hydrogen bonds between POT1 and telomeric DNA. There are many hydrogen bonds between Gln94 and the first guanine of the closest TTAGGG sequence in telomeric single-stranded DNA. These Gln94 and the guanine have a large difference in root mean square fluctuation between single and binding states. We found that Gln94 and guanine are important components of the binding system, and they are related to its stability.
Hattori, Yuya; Yokoya, Akinari; Watanabe, Ritsuko
BMC Systems Biology (Internet), 9, p.90_1 - 90_22, 2015/12
Times Cited Count:17 Percentile:66.49(Mathematical & Computational Biology)The radiation-induced bystander effect is a biological response observed in non-irradiated cells surrounding an irradiated cell, which is known to be caused by two intercellular signaling pathways. However, the behavior of the signals is largely unknown. To investigate the role of these signaling pathways, we developed a mathematical model to describe the cellular response to direct irradiation and the bystander effect, with a particular focus on cell-cycle modification. The analysis of model dynamics revealed that bystander effect on cell cycle modification was different between low-dose irradiation and high-dose irradiation. We demonstrated that signaling through both pathways induced cell cycle modification via the bystander effect. By simulating various special and temporal conditions of irradiation and cell characteristics, our model will be a powerful tool for the analysis of the bystander effect.